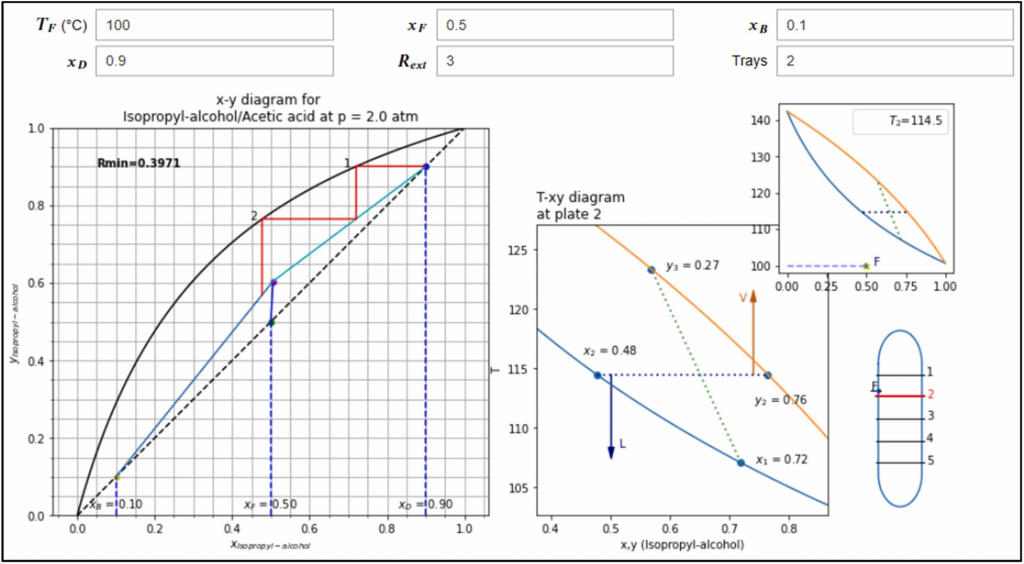
A new article has been published in the journal Education for Chemical Engineers. In the paper, we develop interactive Jupyter Notebooks (JNs) to support Chemical Engineering modules about balances, reactors and fluid separation, and prove that students improve their learning them. You can read the paper here, and check the JNs in the github repository.